Data Cleaning with Awk
I use Python for most of my machine learning and data science work but I find the Unix command line tools, and Awk in particular, super useful for interactively processing and formatting data as a preliminary step before continuing the analysis in Python. I've been using it for basic data cleaning, filtering, merging, reformatting, and data transformation tasks. Here are some reasons to use the command line for preprocessing data:
- Well known. These tools are well known and they exist on pretty much every Unix-like operating system. You can easily share a simple preprocessing pipeline as a shell script with someone and they can run it without installing any dependencies.
- They're pretty fast. These tools were designed to work with streams of text so you can work through large CSV or plain text files without worrying too much about speed, especially if you have a modern machine.
- Concise syntax leads to easy interactive exploration of the data. You don't have to type a lot of boilerplate to get things done.
- Easily composable. The Unix philosophy of using combining simple programs to do complex tasks makes it easy to get stuff done quickly. The CLI allows you to create simple data processing pipelines without writing boilerplate code. After iteratively (and interactively) building up a pipeline on the command line, you can create a simple shell script so that you can re-use it later on new data.
This post goes over how to do some basic data cleaning tasks on a log file from a sound monitoring device. It was created to debug some sound level monitoring issues and it was captured from a terminal emulation application using a manual process and has some formatting issues. I'm going to inspect the log and clean it up so that I can bring it into Python for a deeper analysis.
Determining the number of lines #
The file is called debug.log. Let's see how many lines are in the file:
wc -l debug.log 4266 debug.logThe wc command counts the number of lines, words, and characters by default but we are passing -l so that we only get the number of lines. This log is pretty small, only containing 4266 lines.
Inspecting the log file #
Next, let's take a look at the first 10 lines and the last 10 lines.
(head && echo "..." && tail) < debug.log[2023-09-14 10:53:12.517] RolledUp Spl: 9193.21 Rolledup SplDb: 39.63
[2023-09-14 10:53:12.517] Raw Spl: 8696.51 Raw SplDb: 39.39 RolledUp Spl: 9189.56 Rolledup SplDb: 39.63
[2023-09-14 10:53:12.517] Raw Spl: 11274.82 Raw SplDb: 40.52 RolledUp Spl: 9204.87 Rolledup SplDb: 39.64
[2023-09-14 10:53:12.534] Raw Spl: 8856.91 Raw SplDb: 39.47 RolledUp Spl: 9202.32 Rolledup SplDb: 39.64
[2023-09-14 10:53:12.534] Raw Spl: 8269.93 Raw SplDb: 39.18 RolledUp Spl: 9195.47 Rolledup SplDb: 39.64
[2023-09-14 10:53:12.534] Raw Spl: 7676.67 Raw SplDb: 38.85 RolledUp Spl: 9184.32 Rolledup SplDb: 39.63
[2023-09-14 10:53:12.534] Raw Spl: 9341.36 Raw SplDb: 39.70 RolledUp Spl: 9185.47 Rolledup SplDb: 39.63
[2023-09-14 10:53:12.534] Raw Spl: 7895.02 Raw SplDb: 38.97 RolledUp Spl: 9176.00 Rolledup SplDb: 39.63
[2023-09-14 10:53:12.534] Raw Spl: 7766.24 Raw SplDb: 38.90 RolledUp Spl: 9165.65 Rolledup SplDb: 39.62
...
[2023-09-14 10:54:26.219] Raw Spl: 8466.11 Raw SplDb: 39.28 RolledUp Spl: 9572.91 Rolledup SplDb: 39.81
[2023-09-14 10:54:26.219] Raw Spl: 7873.84 Raw SplDb: 38.96 RolledUp Spl: 9560.43 Rolledup SplDb: 39.80
[2023-09-14 10:54:26.256] Raw Spl: 7861.70 Raw SplDb: 38.96 RolledUp Spl: 9547.96 Rolledup SplDb: 39.80
[2023-09-14 10:54:26.256] Raw Spl: 7918.97 Raw SplDb: 38.99 RolledUp Spl: 9536.00 Rolledup SplDb: 39.79
[2023-09-14 10:54:26.286] Raw Spl: 8022.28 Raw SplDb: 39.04 RolledUp Spl: 9524.89 Rolledup SplDb: 39.79
[2023-09-14 10:54:26.321] Raw Spl: 9248.24 Raw SplDb: 39.66 RolledUp Spl: 9522.85 Rolledup SplDb: 39.79
[2023-09-14 10:54:26.321] Raw Spl: 10626.55 Raw SplDb: 40.26 RolledUp Spl: 9530.96 Rolledup SplDb: 39.79
[2023-09-14 10:54:26.321] Raw Spl: 10752.65 Raw SplDb: 40.32 RolledUp Spl: 9539.93 Rolledup SplDb: 39.80
[2023-09-14 10:54:26.357] Raw Spl: 9736.78 Raw SplDb: 39.88 RolledUp Spl: 9541.38 Rolledup SplDb: 39.80
[2023-09-14 10:54:26.357] Raw Spl: 8368.17 Raw SplDb: 39.23Step-by-step breakdown (click to hide)
head |
| Print out the first 10 lines of the input. If you want to choose the number of lines printed, you can use the -n parameter. For example, head -n 30 will print the first 30 lines of the file. |
&& |
| Run commands sequence, but only if the previous command succeeded without returning an error. For example A && B calls A and if A returns no error code then B will be called |
echo "..." |
| The echo command just prints a string. In this case I'm printing "..." just to separate the beginning lines from the ending lines. |
tail |
| Print out the last 10 lines of the input |
( ) (parentheses) |
| Group the commands together, allowing the head, echo, and tail commands to operate on a single input stream. |
< debug.log (file redirection) |
| part reads the debug.log file and feeds it as standard input to the group of commands in parentheses. |
It looks like there are some blank lines and some lines where the number of fields are inconsistent. Inconsistent formatting is extremely common when dealing with data. Let's see how we can clean up the formatting using a tool called Awk.
Quick intro to Awk #
AWK is a tool and programming language used to process text streams. It's easy to understand how it works using an example. Say we have a text file, emp.data, that contains employee data with names, pay rates, and number of hours worked:
Beth 21 0
Dan 19 0
Kathy 15.50 10
Mark 25 20
Mary 22.50 22
Susie 17 18The goal is to get the name and total pay for each employee that worked more than zero hours. This can be done using the following Awk program:
awk '$3 > 0 { print $1, $2 * $3 }' emp.dataKathy 155
Mark 500
Mary 495
Susie 306The structure of the above command is like this:
awk 'AWK_PROGRAM' inputfileAnd an Awk program itself (the part in single quotes) has the following simple structure:
pattern1 { action1 } pattern2 { action2 }So in the program above, I'm using the pattern $3 > 0 to select all lines where total hours worked (3rd field) is greater than zero.
In the action part, print $1, $2 * $3, I'm just printing the first and product of the 2nd and 3rd fields. Awk is incredibly useful for wrangling text data. By default, Awk uses spaces as the field separator.
You can also operate on multiple files:
awk 'AWK_PROGRAM' inputfile1 inputfile2AWK allows you to use regular expressions for the pattern, such as /warning/ to match all lines that contain the word warning or !/warning/ to match all lines that do not contain the word warning.
You can also look for match on a given field, using regular expressions, or by checking for an exact match:
$1 == "warning" (first field is exactly warning)
$1 ~ "warning" (first field contains warning).
There are also special pattern blocks. The BEGIN pattern is executed once before any input lines are read. The END pattern executes once after all input lines have been read.
The Awk command optionally allows you to pass a program contained in another file using the -f option:
awk -f my_program inputfile
And, of course, it can be chained using pipes to work on standard input. Here's a program to find the average size (in bytes) of all files and directories in the current directory:
ls -l | awk '$1 != "total" {sum += $5; n++} END {print sum/n}'111898Awk has many useful built-in variables:
| Variable | Description | Default |
|---|---|---|
NR |
Number of input records so far (total) | N/A (read only) |
FNR |
Number of input records so far (current file) | N/A (read only) |
NF |
Number of fields in the current record | N/A (read only) |
FS |
Input field separator | Space character |
RS |
Input record separator | New line character |
OFS |
Output field separator | Space character |
ORS |
Output record separator | New line character |
Find the full list of Awk variables here
FS, RS, OFS, and ORS can be set in the BEGIN action. Alternatively, you can use the -F command line option to set the field separator without a using the special BEGIN pattern.
Useful Awk one-liners #
For me, the best Awk programs are short. Here are some useful one liners.
Print the total number of input lines:
awk 'END { print NR }'Print the first 5 lines:
awk 'NR <= 10'Print the total number of lines that contain error:
awk '/error/ {n++} END {print n}'Print every line, with the first field replaced by the line number:
awk '{$1 = NR; print}'Print the sums of the fields of every line:
awk '{sum = 0; for (i=1; i <= NF; i++) sum += $i; print sum;}'Concatenate multiple CSV files and remove all headers:
awk 'FNR > 1' *.csv > merged.csvConcatenate multiple CSV files (keeping the header of the first file):
awk '(NR == 1) || (FNR > 1)' *.csv > merged.csvSome of the above examples are from a book called The AWK Programming Language, written by the authors of Awk. I highly recommend this book. It's a short read but it gives a very clear and practical tutorial on Awk and it's capabilities. Okay, back to the log file.
Determining the expected number of fields #
This command will show us the number of lines for each field count.
awk '{print NF}' debug.log | sort -nr | uniq -c4256 14
1 13
2 8
7 5
1 1Step-by-step breakdown
awk '{print NF}' debug.log |
| For each line, print the number of fields. Since I'm not providing a pattern to match, every line in the input will be processed. NF is an Awk variable that is set to the number of fields in the current line |
| sort -nr |
| The vertical line is the pipe operator which pipes the output of one program to the input of another. In this case, I'm piping the results of the previous Awk command to sort, which sorts the results numerically (-n) in descending (-r) order |
| uniq -c |
| Finally, this part uses the uniq command to deduplicate the lines containing the number of fields and add up the number of instances of each field count. |
So the vast majority of lines have 14 fields. Since there's so few, I'm going to print out the lines which have more or less than 14 fields.
awk 'NF != 14 {print NR ": " $0}' debug.log1:
2: [2023-09-14 10:53:12.517] RolledUp Spl: 9193.21 Rolledup SplDb: 39.63
110: [2023-09-14 10:53:12.653] Raw Spl: 7835.94 Raw SplDb: 38.94 Ro: 8894.31 Rolledup SplDb: 39.49
661: [2023-09-14 10:53:22.214] !! wdtcount 1
1215: [2023-09-14 10:53:32.209] !! wdtcount 1
1764: [2023-09-14 10:53:42.235] !! wdtcount 1
2325: [2023-09-14 10:53:52.266] !! wdtcount 1
2892: [2023-09-14 10:54:02.298] !! wdtcount 1
3465: [2023-09-14 10:54:12.335] !! wdtcount 1
4037: [2023-09-14 10:54:22.331] !! wdtcount 1
4267: [2023-09-14 10:54:26.357] Raw Spl: 8368.17 Raw SplDb: 39.23Dropping badly formatted lines #
Since I'm only interested in the signal data, I can safely drop these lines.
awk 'NF == 14' debug.log[2023-09-14 10:53:12.517] Raw Spl: 8696.51 Raw SplDb: 39.39 RolledUp Spl: 9189.56 Rolledup SplDb: 39.63
[2023-09-14 10:53:12.517] Raw Spl: 11274.82 Raw SplDb: 40.52 RolledUp Spl: 9204.87 Rolledup SplDb: 39.64
[2023-09-14 10:53:12.534] Raw Spl: 8856.91 Raw SplDb: 39.47 RolledUp Spl: 9202.32 Rolledup SplDb: 39.64
[2023-09-14 10:53:12.534] Raw Spl: 8269.93 Raw SplDb: 39.18 RolledUp Spl: 9195.47 Rolledup SplDb: 39.64
[2023-09-14 10:53:12.534] Raw Spl: 7676.67 Raw SplDb: 38.85 RolledUp Spl: 9184.32 Rolledup SplDb: 39.63
[2023-09-14 10:53:12.534] Raw Spl: 9341.36 Raw SplDb: 39.70 RolledUp Spl: 9185.47 Rolledup SplDb: 39.63
[2023-09-14 10:53:12.534] Raw Spl: 7895.02 Raw SplDb: 38.97 RolledUp Spl: 9176.00 Rolledup SplDb: 39.63
[2023-09-14 10:53:12.534] Raw Spl: 7766.24 Raw SplDb: 38.90 RolledUp Spl: 9165.65 Rolledup SplDb: 39.62
[2023-09-14 10:53:12.534] Raw Spl: 10842.39 Raw SplDb: 40.35 RolledUp Spl: 9177.96 Rolledup SplDb: 39.63
[2023-09-14 10:53:12.534] Raw Spl: 16765.23 Raw SplDb: 42.24 RolledUp Spl: 9233.67 Rolledup SplDb: 39.65
...Inspecting the field names #
I want to take a look at the field names to make sure they're consistent. I'll combine the previous Awk program with the following new command which will count the instances of the different field names.
awk 'BEGIN { OFS="\n" } NF == 14 {print $3 $4, $6 $7, $9 $10, $12 $13 }' \
debug.log | sort | tr -d ":" | uniq -c4253 RawSpl
4256 RawSplDb
4256 RolledUpSpl
4256 RolledupSplDb
3 zRawSplStep-by-step breakdown
BEGIN { OFS="\n" } |
| Before processing any lines, set the output field separator to a newline character so that it prints one signal name per line. |
NF == 14 |
| Same as before, only process lines that have exactly 14 fields. |
{ print $3 $4, $6 $7, $9 $10, $12 $13 } |
| This Awk action prints the field names. In the input file, the signal names are separated by space (eg. Raw SPL) so they span 2 fields. When using print in Awk, two variables side by side or with a space in between get concatenated, so $3 $4 results in RawSPL. |
\ |
| The backslash lets you continue the program on the next line, just for readability. |
debug.log |
| debug.log is the name of the input file. |
sort | tr -d ":" | uniq -c |
| Sort the results (in this case, alphabetically), delete colons using the translate command, and then count the number of instances of each field name. |
You can also accomplish the same thing solely using Awk, but it's a bit less clear, in my opinion:
awk 'NF == 14 {counts[$3 $4]+=1; counts[$6 $7]+=1; counts[$9 $10]+=1; counts[$12 $13]+=1} \
END {for (name in counts) print name, counts[name]}' debug.logzRawSpl: 3
RawSplDb: 4256
RolledUpSpl: 4256
RawSpl: 4253
RolledupSplDb: 4256Step-by-step breakdown
NF == 14 |
| Only process lines that have exactly 14 fields |
{ counts[$3 $4]+=1; counts[$6 $7]+=1; counts[$9 $10]+=1; counts[$12 $13]+=1 } |
| This action uses a variable, counts, which holds an associative array (similar to a Python dictionary). By default, Awk uses spaces as the field separator. In the log file, multi-word field names in the input are separated by space so they span 2 fields. In an Awk action, printing different variables without a comma in between results in string concatenation (the space is optional), so c[$3 $4] translates to c[RawSpl] (concatenate the 3rd and 4th fields). In Awk, you don't have to declare a variable before using it. |
END { for (name in c) print name, c[name] } |
| The END here tells Awk to run this part of the program after processing the last line of the input. I'm iterating over the associate array and printing the field names and number of instances of each. |
It looks like the character z is showing up in front of the RawSPL field, but only sometimes. Oddly enough, the presence of the z signifies that an event occurred at that time. Based on this I'd like to do the following:
- The presence of a the letter z is a weird way of encoding events, it would be better to just have a boolean event column that is 1 if the event occurs and 0 otherwise.
- The timestamp field uses square brackets. I'd like to change them to double quotes instead, and replace the single space between the date and time with a T to make it more like a standard ISO date time string that can be easily parsed by pandas.
- Convert the file to CSV format.
Building on our earlier pipeline, here is the new command which accomplishes all of the above.
tr '\r[]' '\n""' < debug.log | \
awk 'BEGIN { OFS="," } \
NF == 14 { event = $3~/z/ ? 1:0; \
print $1 "T" $2, $5, $8, $11, $14, event }' > data.csvHere's what the first 10 lines of the csv file look like:
"2023-09-14T10:53:12.517",8696.51,39.39,9189.56,39.63,0
"2023-09-14T10:53:12.517",11274.82,40.52,9204.87,39.64,0
"2023-09-14T10:53:12.534",8856.91,39.47,9202.32,39.64,0
"2023-09-14T10:53:12.534",8269.93,39.18,9195.47,39.64,0
"2023-09-14T10:53:12.534",7676.67,38.85,9184.32,39.63,0
"2023-09-14T10:53:12.534",9341.36,39.70,9185.47,39.63,0
"2023-09-14T10:53:12.534",7895.02,38.97,9176.00,39.63,0
"2023-09-14T10:53:12.534",7766.24,38.90,9165.65,39.62,0
"2023-09-14T10:53:12.534",10842.39,40.35,9177.96,39.63,0
"2023-09-14T10:53:12.534",16765.23,42.24,9233.67,39.65,0Adding a CSV Header #
It would be nice to prepend a CSV header to our data.csv file. We can generate a header using the names that are already in the file. Here's how to do that, re-using one of our commands from earlier:
awk 'BEGIN { OFS="," } \
NF == 14 { print "Timestamp", $3 $4, $6 $7, $9 $10, $12 $13, "Event" }' \
debug.log | sort | tr -d ":z" | uniq | cat - data.csv > data.formatted.csvTimestamp,RawSpl,RawSplDb,RolledUpSpl,RolledupSplDb,Event
"2023-09-14T10:53:12.517",8696.51,39.39,9189.56,39.63,0
"2023-09-14T10:53:12.517",11274.82,40.52,9204.87,39.64,0
"2023-09-14T10:53:12.534",8856.91,39.47,9202.32,39.64,0
"2023-09-14T10:53:12.534",8269.93,39.18,9195.47,39.64,0
"2023-09-14T10:53:12.534",7676.67,38.85,9184.32,39.63,0
"2023-09-14T10:53:12.534",9341.36,39.70,9185.47,39.63,0
"2023-09-14T10:53:12.534",7895.02,38.97,9176.00,39.63,0
"2023-09-14T10:53:12.534",7766.24,38.90,9165.65,39.62,0
"2023-09-14T10:53:12.534",10842.39,40.35,9177.96,39.63,0Step-by-step breakdown
BEGIN { OFS="," } |
| Before processing any lines, set the output field separator to a comma. |
{print "Timestamp", $3 $4, $6 $7, $9 $10, $12 $13, "Event"}' debug.log |
| Same as an earlier program, concatenate the field names, and prepend and append Timestamp and Event field names respectively. |
| sort | tr -d ":z" | uniq |
| This is a pipeline to get a unique list of field names from the original data file and delete the : and z characters. |
cat - data.csv > data.formatted.csv |
| Prepend the result to the csv file we created earlier. The cat program concatenates text from a sequence of text streams or files. The hyphen, -, tells cat to use standard input for the first file. |
Analyzing with Python #
Now that the data has been cleaned up and formatted correctly, we can import the CSV using pandas:
import pandas as pd
df = pd.read_csv('data.formatted.csv', parse_dates=['Timestamp'])
print(df) Timestamp RawSpl RawSplDb RolledUpSpl RolledupSplDb Event
0 2023-09-14T10:53:12.517 8696.51 39.39 9189.56 39.63 0
1 2023-09-14T10:53:12.517 11274.82 40.52 9204.87 39.64 0
2 2023-09-14T10:53:12.534 8856.91 39.47 9202.32 39.64 0
3 2023-09-14T10:53:12.534 8269.93 39.18 9195.47 39.64 0
4 2023-09-14T10:53:12.534 7676.67 38.85 9184.32 39.63 0
... ... ... ... ... ... ...
4251 2023-09-14T10:54:26.286 8022.28 39.04 9524.89 39.79 0
4252 2023-09-14T10:54:26.321 9248.24 39.66 9522.85 39.79 0
4253 2023-09-14T10:54:26.321 10626.55 40.26 9530.96 39.79 0
4254 2023-09-14T10:54:26.321 10752.65 40.32 9539.93 39.80 0
4255 2023-09-14T10:54:26.357 9736.78 39.88 9541.38 39.80 0You can see that everything was parsed correctly if we print the data types that pandas automatically detected:
df.dtypesTimestamp datetime64[ns]
RawSpl float64
RawSplDb float64
RolledUpSpl float64
RolledupSplDb float64
Event int64
dtype: objectQuick plot of all signals, to get an overview:
from matplotlib import pyplot as plt
plt.style.use('seaborn')
df.set_index('Timestamp').plot(figsize=(12,10),
subplots=True,
sharex=True,
grid=True)
plt.tight_layout()
plt.show()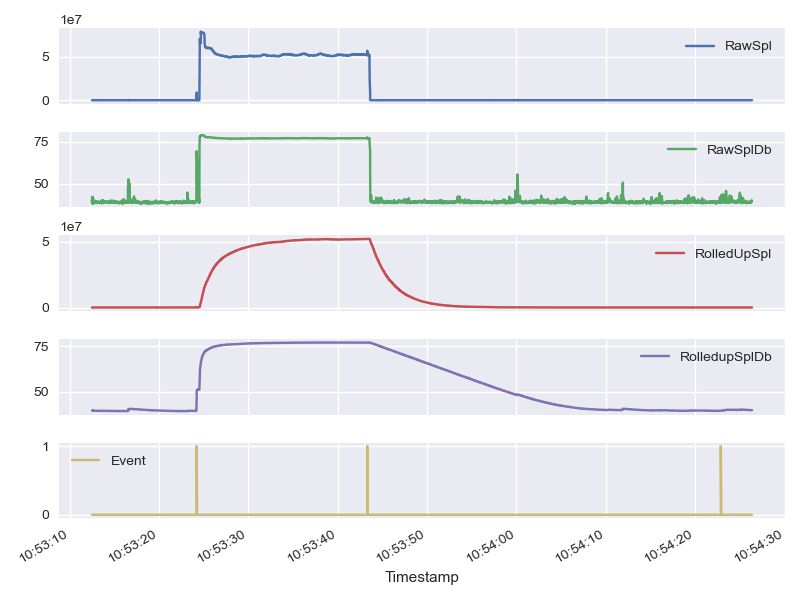 |
|---|
Plotting the 4 signals from the formatted csv file |
Wrap up #
For me, Python is still the best tool for exploratory data analysis but the shell is perfect for simple data cleaning and analysis tasks - especially for preliminary analysis and reformatting tasks like I've shown above. Being able to interactively process large files without writing any boilerplate is convenient and makes for an efficient workflow. You can also use Jupyter's !command feature, which allows you to run shell commands directly from a notebook. At some point, it makes sense to switch to Python and use the full arsenal of numerical libraries for a deeper analysis but for preliminary analysis and data formating/cleaning, the CLI tools are really effective.
- Previous: Batch Converting JPEGs to HEICs